In June, South Korean regulators authorized the first-ever medicine, a COVID-19 vaccine, to be made from a novel protein designed by humans. The vaccine is based on a spherical protein ‘nanoparticle’ that was created by researchers nearly a decade ago, through a labour-intensive trial-and error-process1.
Now, thanks to gargantuan advances in artificial intelligence (AI), a team led by David Baker, a biochemist at the University of Washington (UW) in Seattle, reports in Science2,3 that it can design such molecules in seconds instead of months.
‘The entire protein universe’: AI predicts shape of nearly every known protein
Such efforts are a part of a scientific sea change, as AI tools such as DeepMind’s protein-structure-prediction software AlphaFold are embraced by life scientists. In July, DeepMind revealed that the latest version of AlphaFold had predicted structures for every protein known to science. And recent months have seen an explosive growth in AI tools — some based on AlphaFold — that can quickly dream up completely new proteins. Previously, this had been a painstaking pursuit with high failure rates.
“Since AlphaFold, there’s been a shift in the way we work with protein design,” says Noelia Ferruz, a computational biologist at the University of Girona, Spain. “We are witnessing very exciting times.”
Most efforts are focused on tools that can help to make original proteins, shaped unlike anything in nature, without much focus on what these molecules can do. But researchers — and a growing number of companies that are applying AI to protein design — would like to design proteins that can do useful things, from cleaning up toxic waste to treating diseases. Among the companies that are working towards this goal are DeepMind in London and Meta (formerly Facebook) in Menlo Park, California.
“The methods are already really powerful. They’re going to get more powerful,” says Baker. “The question is what problems are you going to solve with them.”
From scratch
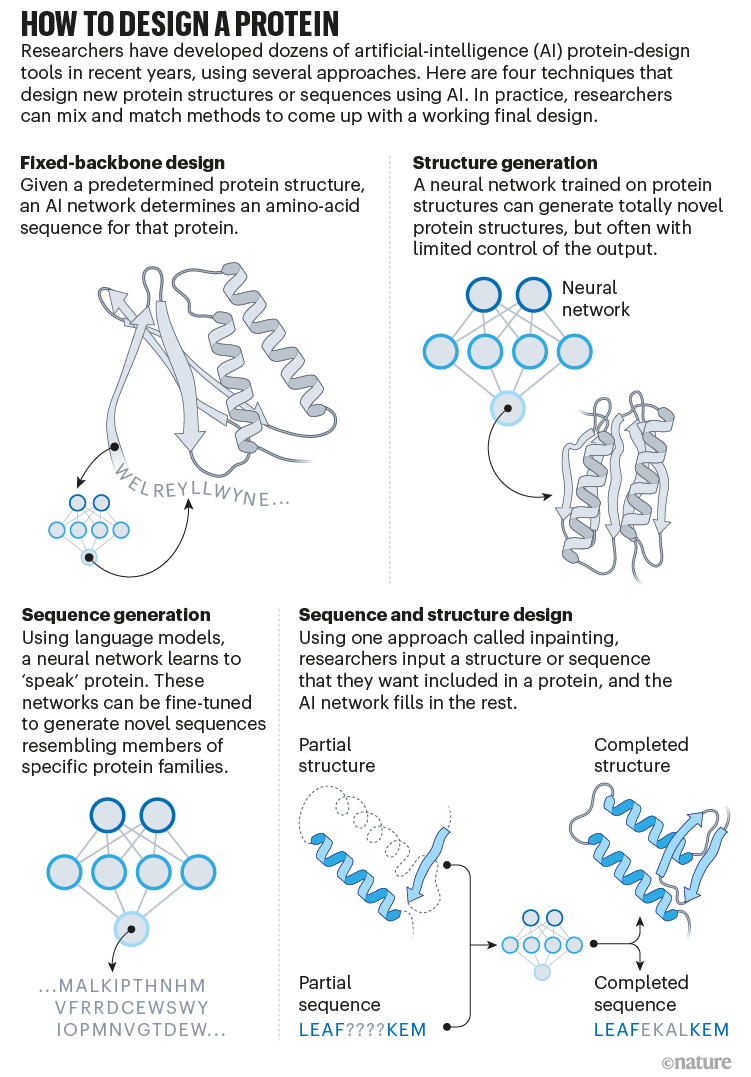
Baker’s laboratory has spent the past three decades making new proteins. Software called Rosetta, which his lab started developing in the 1990s, splits the process into steps. Initially, researchers conceived a shape for a novel protein — often by cobbling together bits of other proteins — and the software deduced a sequence of amino acids that corresponded to this shape.
But these ‘first draft’ proteins rarely folded into the desired shape when made in the lab, and instead ended up stuck in different confirmations. So another step was needed to tweak the protein sequence such that it folded only into a single desired structure. This step, which involved simulating all the ways in which different sequences might fold, was computationally expensive, says Sergey Ovchinnikov, an evolutionary biologist at Harvard University in Cambridge, Massachusetts, who used to work in Baker’s lab. “You would literally have, like, 10,000 computers running for weeks doing this.”
What’s next for AlphaFold and the AI protein-folding revolution
By tweaking AlphaFold and other AI programmes, that time-consuming step has become instantaneous, says Ovchinnikov. In one approach developed by Baker’s team, called hallucination, researchers feed random amino-acid sequences into a structure-prediction network; this alters the structure so that it becomes ever-more protein-like, as judged by the network’s predictions. In a 2021 paper, Baker’s team created more than 100 small, ‘hallucinated’ proteins in the lab and found signs that about one-fifth resembled the predicted shape4.
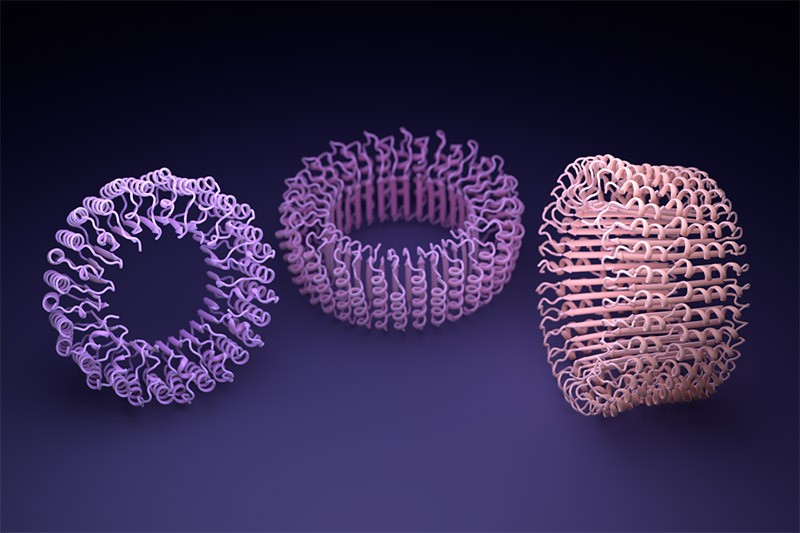
AlphaFold, and a similar tool developed by Baker’s lab called RoseTTAFold, were trained to predict the structure of individual protein chains. But researchers soon discovered that such networks could also model assemblies of multiple interacting proteins. On this basis, Baker and his team were confident they could hallucinate proteins that would self-assemble into nanoparticles of different shapes and sizes; these would be made up of numerous copies of a single protein and would be similar to those on which the COVID-19 vaccine is based.
But when they instructed microorganisms to make their creations in the labs, none of the 150 designs worked. “They didn’t fold at all: they were just gunk at the bottom of the test tube,” says Baker.
Around the same time, another researcher in the lab, machine-learning scientist Justas Dauparas, was developing a deep-learning tool to address what is known as the inverse folding problem — determining a protein sequence that corresponds to a given protein’s overall shape3. The network, called ProteinMPNN, can act as a ‘spellcheck’ for designer proteins created using AlphaFold and other tools, says Ovchinnikov, by tweaking sequences while maintaining the molecules’ overall shape.
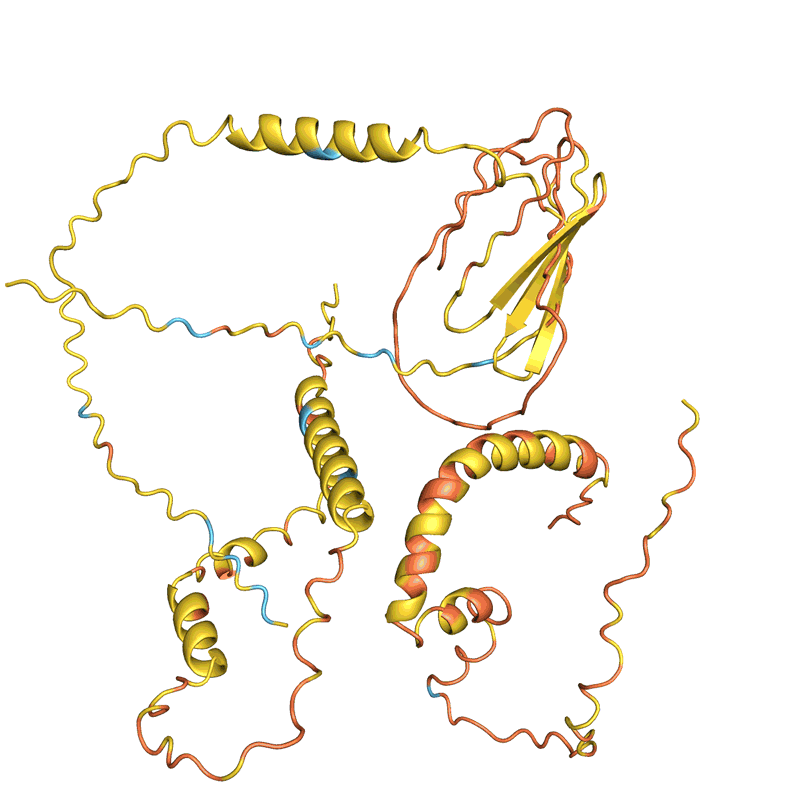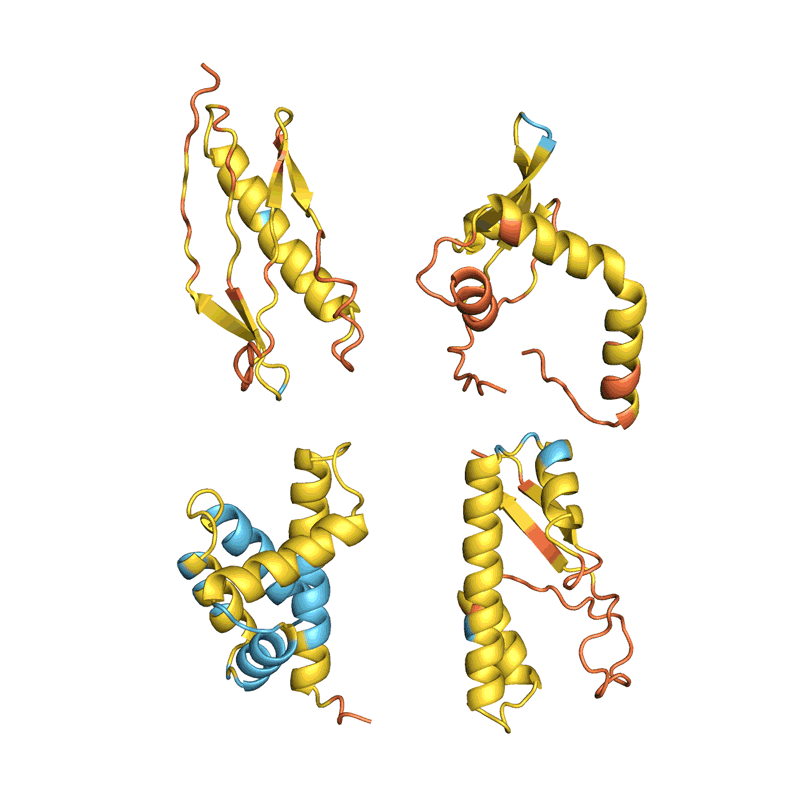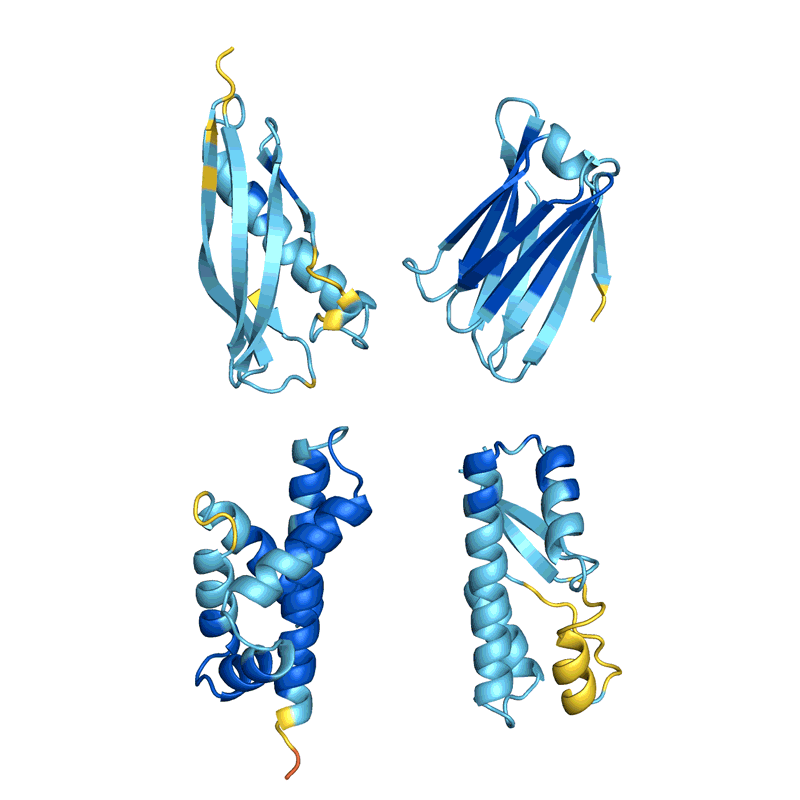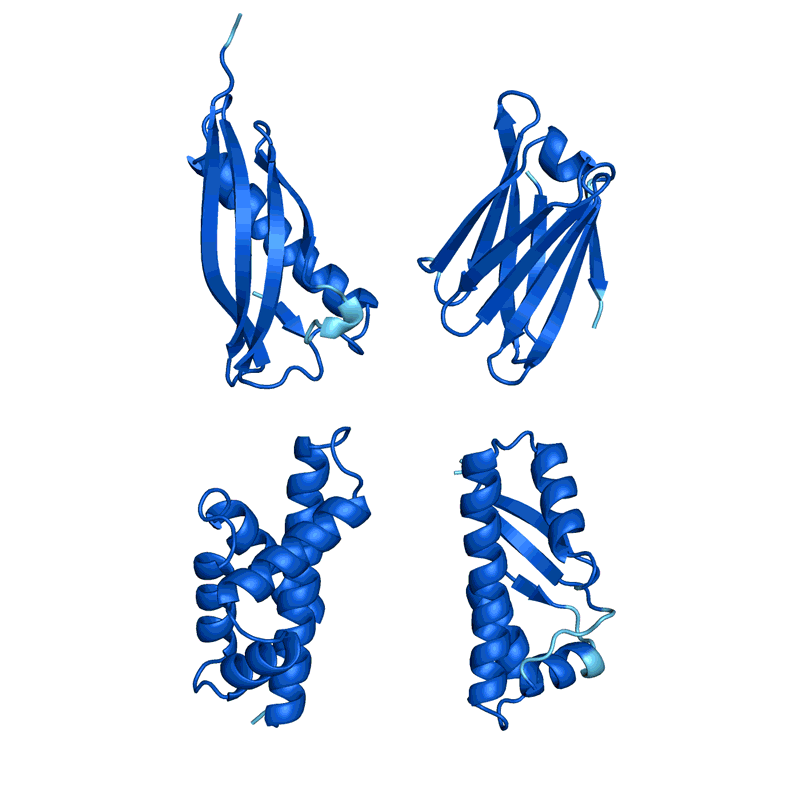
When Baker and his team applied this second network to their hallucinated protein nanoparticles, it had much greater success making the molecules experimentally. The researchers determined the structure of 30 of their new proteins using cryo-electron microscopy and other experimental techniques, and 27 of them matched the AI-led designs2. The team’s creations included giant rings with complex symmetries, unlike anything found in nature. In theory, the approach could be used to design nanoparticles corresponding to almost any symmetric shape, says Lukas Milles, a biophysicist who co-led the effort. “It is electrifying to see what these networks can do.”
Deep-learning revolution
Deep-learning tools such as proteinMPNN have been a game changer in protein design, says Arne Elofsson, a computational biologist at Stockholm University. “You draw your protein, push a button, and you get something that one in ten times works.” Even higher success rates can be achieved by combining multiple neural networks to tackle different parts of the design process, as Baker’s team did in designing the nanoparticles. “Now we have full control over the shape of the protein,” says Ovchinnikov.
Baker’s isn’t the only lab applying AI to protein design. In a review paper posted to the bioRxiv this month, Ferruz and her colleagues counted more than 40 AI protein-design tools that have been developed in recent years, using various approaches5 (see ‘How to design a protein’).
Many of these tools, including proteinMPNN, tackle the inverse folding problem: they specify a sequence that corresponds to a particular structure, often using approaches borrowed from image-recognition tools. Some others are based on an architecture similar to that of language neural networks such as GPT-3, which produces human-like text; but, instead, the tools are capable of producing novel protein sequences. “These networks are able to ‘speak’ proteins,” says Ferruz, who has co-developed one such network6.
With so many protein-design tools available, it’s not always clear how best to compare them, says Chloe Hsu, a machine-learning researcher at the University of California, Berkeley, who developed an inverse folding network with researchers from Meta7.
Many teams gauge their network’s ability to accurately determine the sequence of an existing protein from its structure. But this doesn’t apply for all methods, and it’s not clear how this metric, known as recovery rate, applies to the design of novel proteins, say scientists. Ferruz would like to see a protein-design competition, analogous to the biennial Critical Assessment of protein Structure Prediction (CASP) experiment, in which AlphaFold first demonstrated its superiority over other networks. “It’s a dream. Something like CASP would really move the field forward,” she says.
To the wet lab
Baker and his colleagues are adamant that making a novel protein in the lab is the ultimate test of their methods. Their initial failure to make hallucinated protein assemblies shows this. “AlphaFold thought they were fantastic proteins, but they clearly didn’t work in the wet lab,” says Basile Wicky, a biophysicist in Baker’s lab who co-led the effort, along with Baker, Milles and UW biochemist Alexis Courbet.
But not all scientists developing AI tools for protein design have easy access to experimental set-ups, notes Jinbo Xu, a computational biologist at the Toyota Technological Institute at Chicago in Illinois. Finding a lab to collaborate with can take time, so Xu is establishing his own wet lab to put his team’s creations to the test.
Experiments will also be essential when it comes to designing proteins with specific tasks in mind, says Baker. In July, his team described a pair of AI methods that allow researchers to embed a specific sequence or structure in a novel protein8. They used these approaches to design enzymes that catalyse particular reactions; proteins capable of binding to other molecules; and a protein that could be used in a vaccine against a respiratory virus that is a leading cause of infant hospitalizations.
Last year, DeepMind launched a spin-off company called Isomorphic Labs in London that intends to apply AI tools such as AlphaFold to drug discovery. DeepMind’s chief executive, Demis Hassabis, says that he sees protein design as an obvious and promising application for deep-learning technology, and for AlphaFold in particular. “We’re working quite a lot in the protein design space. It’s pretty early days.”